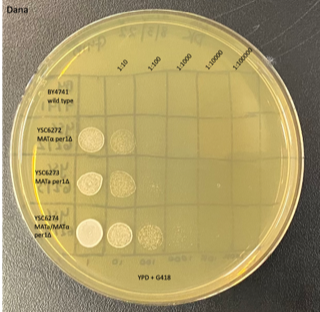
Per1∆ yeast strains have been thawed. The homozygous diploid strain has not been able to grow yet. However both mating types of the haploid deletion strains, as well as the heterozygous diploid strain (one functional copy of Per1), are growing successfully. Encouragingly, both haploid mating types of Per1∆ yeast exhibit significantly attenuated growth compared to the heterozygote with functional Per1 (see attached image). This is an excellent sign that Per1∆ exhibits a rescuable phenotype. Next week the yeast Cure Guides will troubleshoot the growth of the diploid knockout strain and test the sensitivity of the Per1 deletion strains to heat stress, which the literature indicates will further widen the gap between Per1∆ and Per1 +/- growth rates. In parallel, we’re in discussion with UCSF’s small molecule discovery center to ensure that your screens are charged at the external non-profit rate (as opposed to the external for-profit rate, which is higher). I’ll also be meeting with the yeast Cure Guides on Wednesday morning next week, so if you have any specific questions for them, please do share them with me and I will pass them along. Have a great weekend! Justin
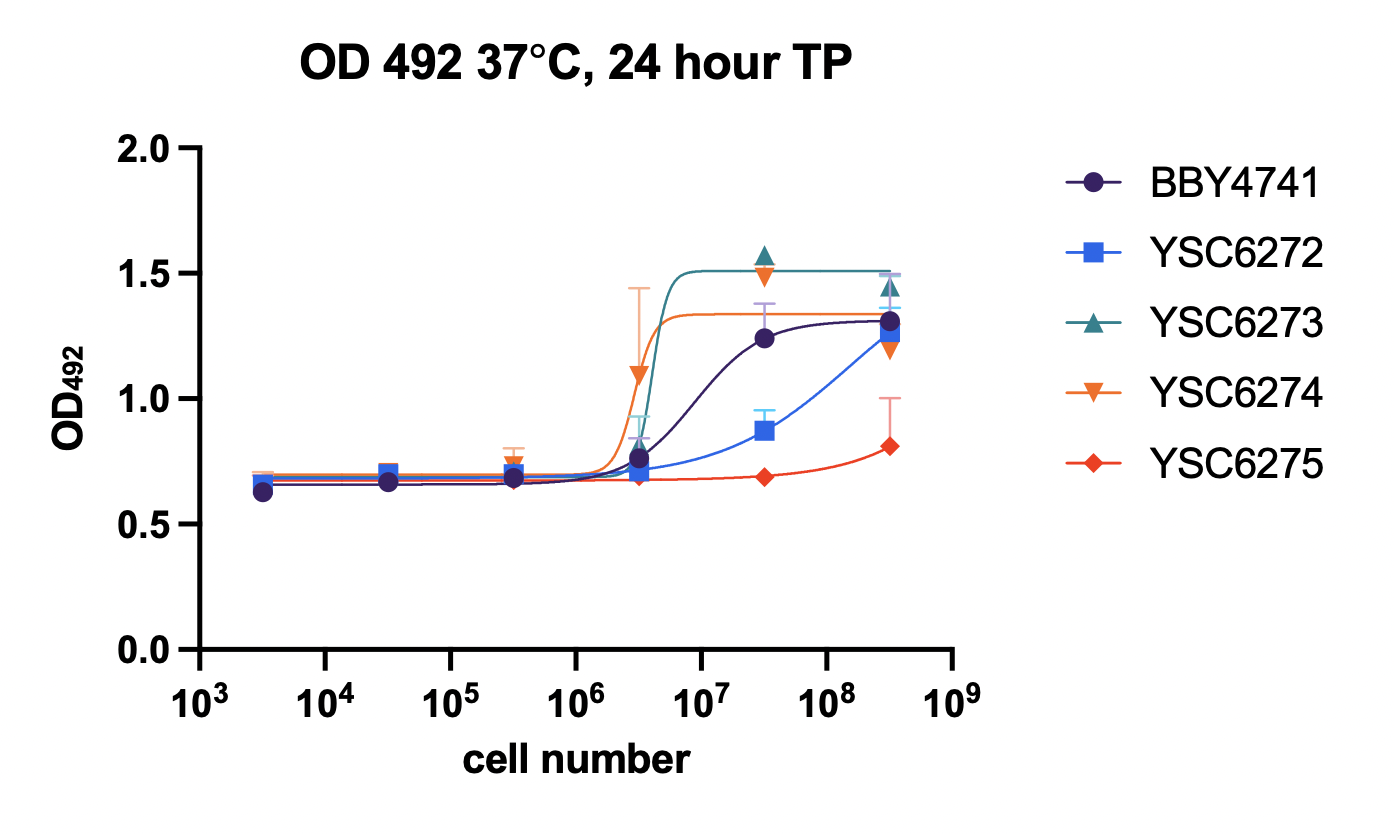
To date, we have successfully validated a growth defect in Per1∆ yeast at 30ºC and 37ºC. This growth rate was measured using two orthogonal methods (OD and the BacTiter-Glo luminescence assay). The X-axis represents the initial cell count, and the Y-axis represents the initial output. See below for a legend identifying the different strains: BBY4741: Wild-type yeast (baseline) YSC6272: MATalpha Per1∆ (deficient) YSC6273: MATa Per1∆ (deficient) YSC6274: Heterozygous diploid Per1∆ (control, one functional Per1 copy) YSC6275: Homozygous diploid Per1∆ (deficient) It is encouraging to see the growth defect even under unstressed conditions (30ºC). With the validated strains and assay in-hand, we are on track to begin screening in late August. Additionally, last week I told you that we were working with UCSF's core facility to be billed at the external non-profit rate for the screen. I'm encouraged to report that we received confirmation that we will be charged at this rate. We will begin setting up a contract on Monday. Lastly, on the Lucy avatar front, we completed sequence drafting earlier today. These sequences will be used to generate mutant yeast. Once we confirm them internally, we will place an order to have them synthesized. Twist estimates that we will receive these sequences before Labor Day, but in my experience it's best to expect that they may take a little longer, as things can go wrong in the synthesis process (up to ~1 additional week or so). Our goal is to be ready to screen these lines by mid-September, but this will be somewhat dependent on Twist. I will keep you posted as things progress here. Let me know if you'd like to discuss the data any further! Have a great weekend, Justin

To date, we have successfully validated a growth defect in Per1∆ yeast at 30ºC and 37ºC. This growth rate was measured using two orthogonal methods (OD and the BacTiter-Glo luminescence assay). The X-axis represents the initial cell count, and the Y-axis represents the initial output. See below for a legend identifying the different strains: BBY4741: Wild-type yeast (baseline) YSC6272: MATalpha Per1∆ (deficient) YSC6273: MATa Per1∆ (deficient) YSC6274: Heterozygous diploid Per1∆ (control, one functional Per1 copy) YSC6275: Homozygous diploid Per1∆ (deficient) It is encouraging to see the growth defect even under unstressed conditions (30ºC). With the validated strains and assay in-hand, we are on track to begin screening in late August. Additionally, last week I told you that we were working with UCSF's core facility to be billed at the external non-profit rate for the screen. I'm encouraged to report that we received confirmation that we will be charged at this rate. We will begin setting up a contract on Monday. Lastly, on the Lucy avatar front, we completed sequence drafting earlier today. These sequences will be used to generate mutant yeast. Once we confirm them internally, we will place an order to have them synthesized. Twist estimates that we will receive these sequences before Labor Day, but in my experience it's best to expect that they may take a little longer, as things can go wrong in the synthesis process (up to ~1 additional week or so). Our goal is to be ready to screen these lines by mid-September, but this will be somewhat dependent on Twist. I will keep you posted as things progress here. Let me know if you'd like to discuss the data any further! Have a great weekend, Justin
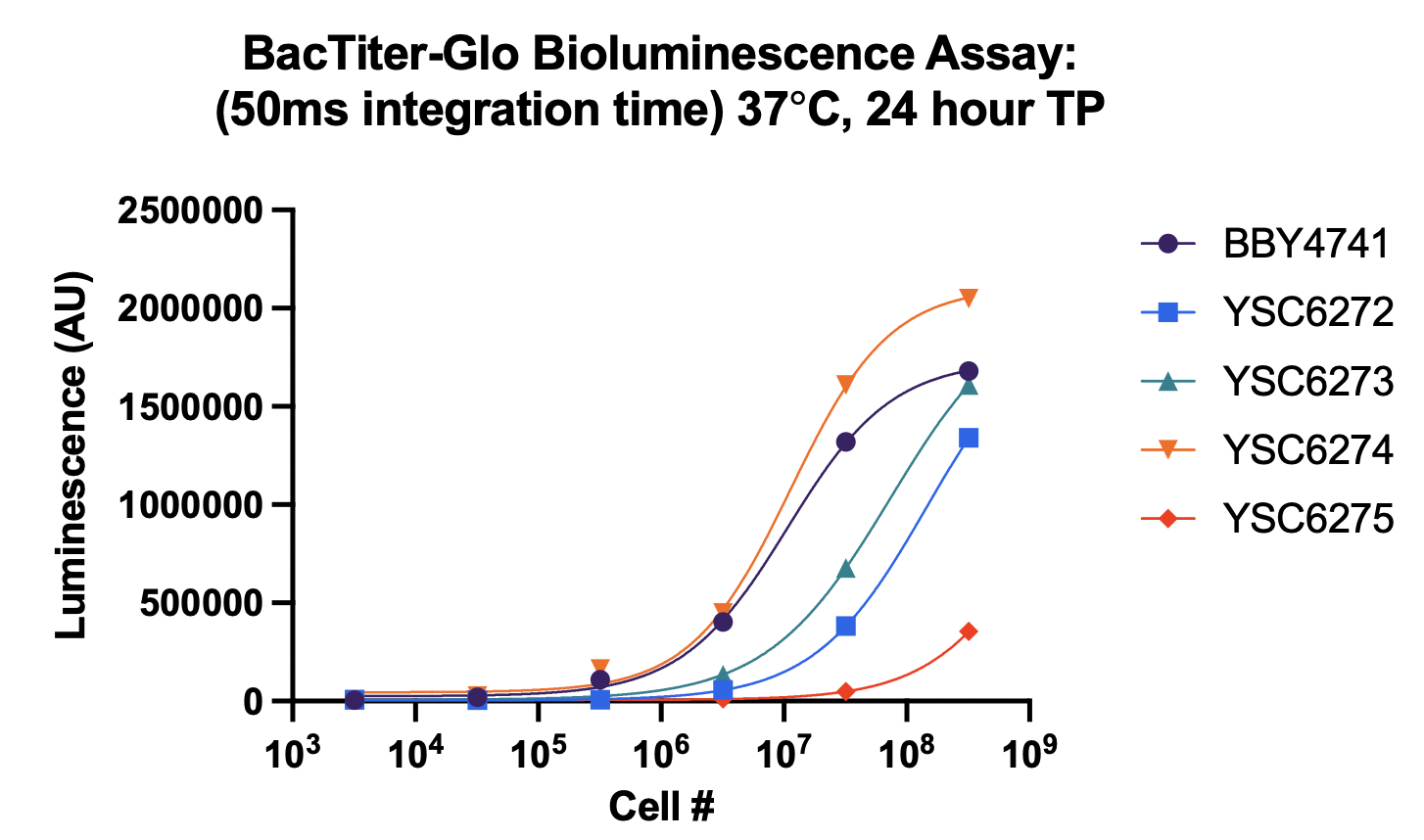
To date, we have successfully validated a growth defect in Per1∆ yeast at 30ºC and 37ºC. This growth rate was measured using two orthogonal methods (OD and the BacTiter-Glo luminescence assay). The X-axis represents the initial cell count, and the Y-axis represents the initial output. See below for a legend identifying the different strains: BBY4741: Wild-type yeast (baseline) YSC6272: MATalpha Per1∆ (deficient) YSC6273: MATa Per1∆ (deficient) YSC6274: Heterozygous diploid Per1∆ (control, one functional Per1 copy) YSC6275: Homozygous diploid Per1∆ (deficient) It is encouraging to see the growth defect even under unstressed conditions (30ºC). With the validated strains and assay in-hand, we are on track to begin screening in late August. Additionally, last week I told you that we were working with UCSF's core facility to be billed at the external non-profit rate for the screen. I'm encouraged to report that we received confirmation that we will be charged at this rate. We will begin setting up a contract on Monday. Lastly, on the Lucy avatar front, we completed sequence drafting earlier today. These sequences will be used to generate mutant yeast. Once we confirm them internally, we will place an order to have them synthesized. Twist estimates that we will receive these sequences before Labor Day, but in my experience it's best to expect that they may take a little longer, as things can go wrong in the synthesis process (up to ~1 additional week or so). Our goal is to be ready to screen these lines by mid-September, but this will be somewhat dependent on Twist. I will keep you posted as things progress here. Let me know if you'd like to discuss the data any further! Have a great weekend, Justin
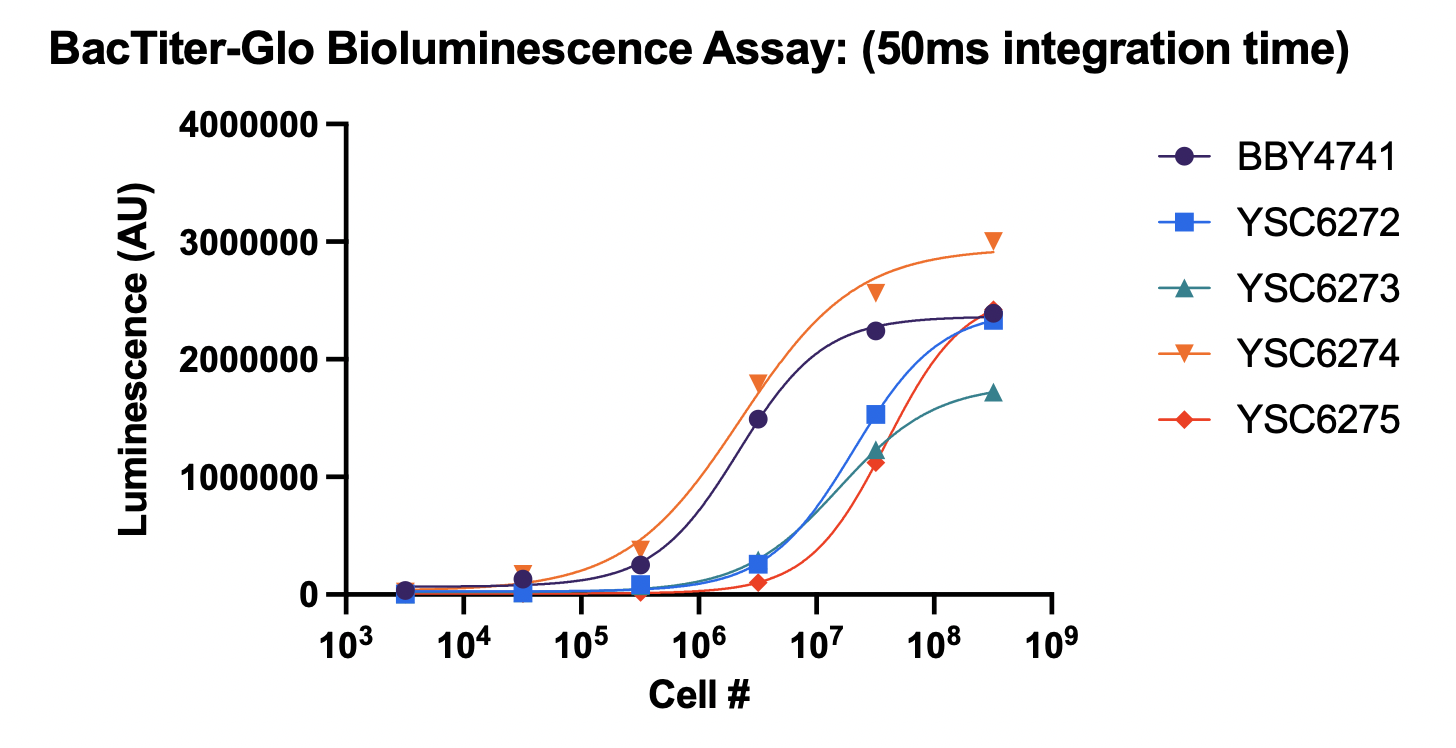
To date, we have successfully validated a growth defect in Per1∆ yeast at 30ºC and 37ºC. This growth rate was measured using two orthogonal methods (OD and the BacTiter-Glo luminescence assay). The X-axis represents the initial cell count, and the Y-axis represents the initial output. See below for a legend identifying the different strains: BBY4741: Wild-type yeast (baseline) YSC6272: MATalpha Per1∆ (deficient) YSC6273: MATa Per1∆ (deficient) YSC6274: Heterozygous diploid Per1∆ (control, one functional Per1 copy) YSC6275: Homozygous diploid Per1∆ (deficient) It is encouraging to see the growth defect even under unstressed conditions (30ºC). With the validated strains and assay in-hand, we are on track to begin screening in late August. Additionally, last week I told you that we were working with UCSF's core facility to be billed at the external non-profit rate for the screen. I'm encouraged to report that we received confirmation that we will be charged at this rate. We will begin setting up a contract on Monday. Lastly, on the Lucy avatar front, we completed sequence drafting earlier today. These sequences will be used to generate mutant yeast. Once we confirm them internally, we will place an order to have them synthesized. Twist estimates that we will receive these sequences before Labor Day, but in my experience it's best to expect that they may take a little longer, as things can go wrong in the synthesis process (up to ~1 additional week or so). Our goal is to be ready to screen these lines by mid-September, but this will be somewhat dependent on Twist. I will keep you posted as things progress here. Let me know if you'd like to discuss the data any further! Have a great weekend, Justin
